NIMBioS Tuesday Seminar Series
In conjunction with the interdisciplinary activities of the National Institute for Mathematical and Biological Synthesis (NIMBioS), a seminar series will be hosted at NIMBioS every other Tuesday at 3:30 p.m. in the NIMBioS Lecture Hall on the 4th floor of 1534 White Ave., Suite 400 Seminar speakers will focus on their research initiatives at the interface of mathematics and many areas of the life sciences. Light refreshments will be served starting at 3 p.m.
Time/Date:
Tuesday, April 5, 2011. 3:30 p.m.*
Location:
Room 403, Blount Hall, 1534 White Ave., Suite 400
Speaker:
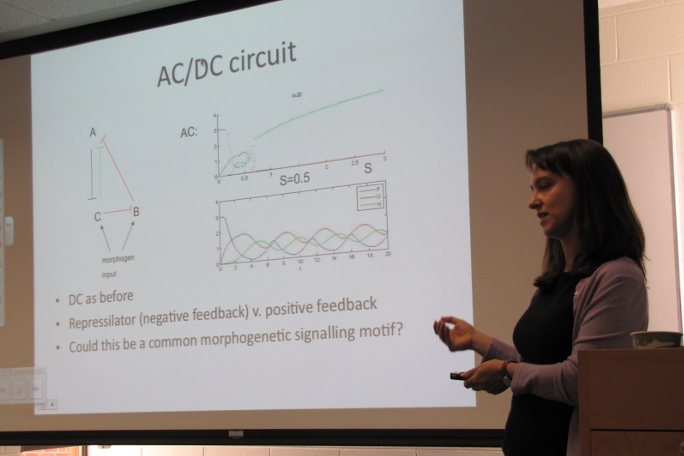
Dr. Karen M. Page, NIMBioS Sabbatical Fellow, Dept. of Mathematics, University College London
Topic: Developmental and evolutionary dynamics
Abstract:
During the course of embryonic development, an initially homogeneous population of cells organizes into an exquisitely patterned organism, consisting of multiple cell types. How the information encoded in the DNA is interpreted to create this 3-dimensional pattern of cell types has intrigued scientists for many years. One strategy for specifying cellular differentiation is the local production and subsequent diffusion of a "morphogen." The signal conferred to cells varies in space and is used by them to decide their fates. An example morphogen is Sonic Hedgehog, which, in vertebrates, specifies neural progenitor domains. We present a mathematical model of the gene regulatory circuit that interprets the Sonic Hedghog signal at the cellular level. We show that the circuit responds to both the level and duration of the signal and confers properties of hysteresis and robustness on cells. We show that, in addition to switch-like behavior, the circuit can also exhibit oscillations. We therefore term the circuit the "AC-DC motif." We suggest that through changes in for example a binding affinity, the motif could have been re-used during the course of evolution for either switch-like or oscillatory functions, both of which are important in embryonic patterning. I will discuss future work which is planned to look at the effects of noise in this neuronal patterning. If there is time, I will also present preliminary work on the migration of neural crest cells and work on evolutionary dynamics. The latter ties together various deterministic mathematical frameworks for describing evolutionary change and I will also outline plans for a stochastic description.
*Join us for refreshments in the NIMBioS Lobby on the 4th floor at 3 p.m.
Seminar Flyer (pdf)
NIMBioS
1122 Volunteer Blvd., Suite 106
University of Tennessee
Knoxville,
TN 37996-3410
PH: (865) 974-9334
FAX: (865) 974-9461
Contact NIMBioS


