| Description | Participants | Agenda | Products | Talks | Posters | Videos |
|---|
Posters for NIMBioS Investigative Workshop
Systems and Synthetic Microbiology
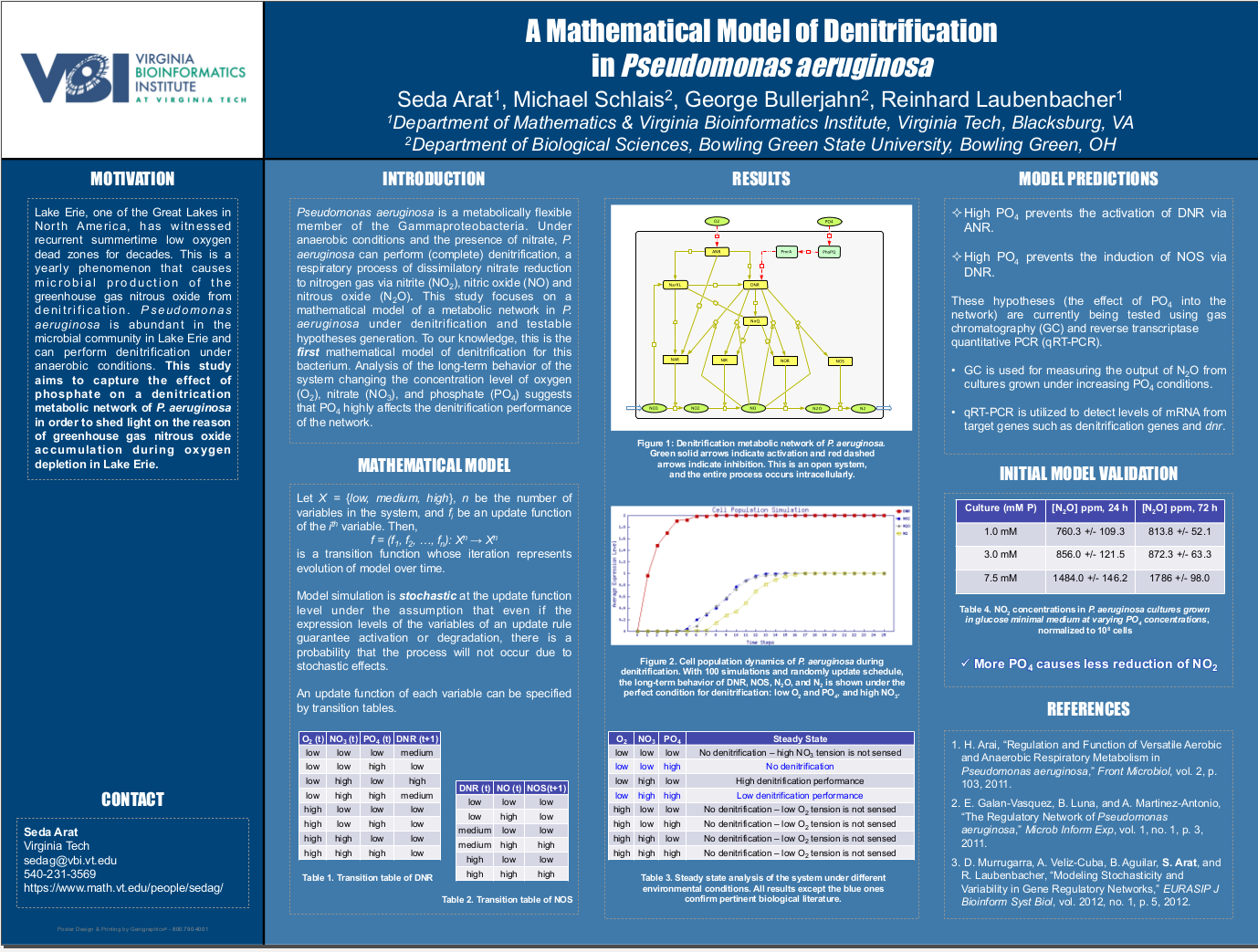 Authors:
Seda Arat1, Michael Schlais2, George Bullerjahn2, Reinhard Laubenbacher1
Authors:
Seda Arat1, Michael Schlais2, George Bullerjahn2, Reinhard Laubenbacher1
Institutions:
(1) Dept. of Mathematics & Virginia Bioinformatics Institute, Virginia Tech, Blacksburg, VA;
(2) Dept. of Biological Sciences, Bowling Green State Univ., Bowling Green, OH
Title:
A mathematical model of denitrification in Pseudomonas aeruginosa.
Abstract:
Pseudomonas aeruginosa is a metabolically
exible member of the Gammaproteobacteria. Under anaerobic conditions and the presence of nitrate, P. aeruginosa can perform (complete) denitrification, a respiratory process of dissimilatory nitrate reduction to nitrogen gas via nitrite (NO2), nitric oxide (NO) and nitrous oxide (N2O). This study focuses
on a mathematical model of a metabolic network in P. aeruginosa under denitrification and
testable hypotheses generation. To our knowledge, this is the first mathematical model of
denitrification for this bacterium. Analysis of the long-term behavior of the system changing the concentration level of oxygen (O2), nitrate (NO3), and phosphate (PO4) suggests
that PO4 highly acts the denitrification performance of the network. This study aims
to capture the ect of phosphate on a denitrification metabolic network of P. aeruginosa
incorporating systems biology approach in order to shed light on the reason of greenhouse
gas N2O accumulation during oxygen depletion in Lake Erie, OH.
Author: Munehiro Asally
Institution: Univ. of California, San Diego
Title: Localized cell death focuses mechanical forces during biofilm formation
Abstract:
From microbial biofilm communities to multicellular organisms, three-dimensional (3D) macroscopic structures develop through a poorly understood interplay between cellular processes and mechanical forces. Investigating wrinkled biofilms of Bacillus subtilis, we discovered a pattern of localized cell death that spatially focuses mechanical forces, and thereby initiates wrinkle formation. Deletion of genes implicated in biofilm development, together with mathematical modeling, revealed that extracellular matrix production underlies the localization of cell death. Simultaneously with cell death, we quantitatively measured mechanical stiffness and movement in wild-type and mutant biofilms. Results suggest that localized cell death provides an outlet for lateral compressive forces, thereby promoting vertical mechanical buckling, which subsequently leads to wrinkle formation. Guided by these findings, we were able to generate artificial wrinkle patterns within biofilms. Formation of 3D structures facilitated by cell death, may underlie self-organization in other developmental systems, and could enable engineering of macroscopic structures from cell populations.
Authors:
Dong-Hoon Chung1,2, Åke Västermark3, Jeremy V. Camp1, Carl Bruder2, Susanna K. Remold4, Yong-Kyu Chu2, Punya Mardhanan2, Ryan McAllister5, and Colleen B. Jonsson1,2,5
Institutions:
(1) Department of Microbiology and Immunology, Univ. of Louisville, Louisville, Kentucky;
(2) Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases, Univ. of Louisville, Louisville, Kentucky;
(3) Institutionen för Neurovetenskap, BMC, Box 593, 751 24 Uppsala, Sweden;
(4) Department of Biology, Univ. of Louisville, Louisville, Kentucky;
(5) Department of Pharmacology and Toxicology, Univ. of Louisville, Louisville, Kentucky
Title:
The murine model for Hantaan virus-induced lethal disease shows two distinct paths in viral evolutionary trajectory with or without ribavirin treatment
Abstract:
In vitro, ribavirin acts as a lethal mutagen in Hantaan virus (HTNV)-infected Vero E6 cells resulting in increased mutation load and viral population extinction. Herein, we asked whether ribavirin treatment in the lethal, suckling mouse model of HTNV would act similarly. HTNV genomic RNA (vRNA) copy number and infectious virus were measured in lungs of mice in untreated and ribavirin-treated mice. In untreated, HTNV-infected mice, vRNA increased to 10 days post-infection (DPI) and thereafter remained constant to 26 DPI. Surprisingly, in ribavirin-treated, HTNV-infected mice, vRNA levels were similar to untreated between 10 and 26 DPI. Infectious virus levels, however, differed; in ribavirin-treated mice, infectious HTNV was significantly decreased relative to untreated mice, suggesting that ribavirin reduced the fitness of the virus (infectious virus produced per vRNA copy). Mutational analysis revealed a similar ribavirin-associated elevation in mutation frequency in HTNV vRNA as was previously reported in vitro. Codon-based analyses of rates of nonsynonymous (dN) and synonymous (dS) substitutions in the S-segment revealed a positive selection on amino acids in the HTNV N protein in the ribavirin-treated vRNA population. In contrast, the vRNA population in untreated HTNV-infected mice showed a lower diversity reflecting purifying selection for the wild-type genome. In summary, these experiments show two different evolutionary paths that Hantavirus may take during infection of a lethal murine model of disease, and the importance of the in vivo host environment in the evolution of the virus that is not apparent in our prior in vitro model system.
Authors:
Amiyaal Ilany1, Adi Barocas1,2, Lee Koren1,3, Michael Kam4 and Eli Geffen1
Institutions:
(1) Dept. of Zoology, Tel Aviv Univ., Tel Aviv, Israel;
(2) Dept. of Zoology and Physiology and Program in Ecology, Univ. of Wyoming, Laramie, WY, USA;
(3) Dept. of Comparative Biology & Experimental Medicine, Faculty of Veterinary Medicine, Univ. of Calgary, Calgary, Canada;
(4) Desert Animal Adaptations and Husbandry, Wyler Dept. of Dryland Agriculture, The Jacob Blaustein Institutes for Desert Research, Ben Gurion Univ. of the Negev, Beer Sheva, Israel.
Title:
The enemy of my enemy is my friend: structural balance in the social networks of a wild mammal
Abstract:
Our understanding of the forces stabilizing specific social structures in animals is limited. The social structure of a population is based on individual social associations, which can be described using network patterns (motifs). Structural balance theory was proposed for exploring social alliances, and suggested that some network motifs are more stable than others in a society. The theory models the presence of specific triads in the network and their effect on the global population structure, based on the differential stability of specific triad configurations. While structural balance was recently shown in human social networks, the theory has never been tested in animal societies. Here we use empirical data from an animal social network to examine, for the first time, the presence and effects of structural balance. We confirm the presence of structural balance in a wild rock hyrax (Procavia capensis) population and show its ability to predict social changes resulting from local instability. We show that new individuals entering the population introduce social instability, which counters the tendency of social relationships to seek balanced structures. Our findings imply that structural balance has a role in the evolution of social structure.
Author:
Tatiana Karpinets
Institution:
Biosciences Division, Oak Ridge National Laboratory
Title: Revealing regularities in large datasets with biological annotations using networks and rules of their association
Abstract:
The expanse of experimental data produced by modern biotechnologies requires the development and application of novel computational tools for their processing. Combined in one table this mash up of qualitative and quantitative data is challenging to analyze and interpret. To address this challenge we developed a computational framework that expands current approaches to viewing, visualizing, searching, and analyzing information in large databases with biological annotations. The approach is based on two novel concepts, the type-value format and the association network (Anet) that supplement the idea of association rules (Arules) produced by 'Apriori'. Type-value format simplifies computational processing, filtering and grouping of biological annotations by preserving their two-level structural organization in the transaction records and further in Arules and Anets. For annotation types presented by quantitative data, such as genome size or GC content, quantities are replaced with their quality levels based on distributions of the quantities in the dataset records. Association network provides a way to link a diverse set of annotations directly, by the number of transactions where each pair of annotations co-occurred, and indirectly, by considering similarity between profiles of their co-occurrences with other annotations. Monte Carlo simulation is used to assess the significance of similarity by p-value. The resulting Anet is further analyzed and visualized at different levels of resolution, or p-value thresholds, using clustering and visualization techniques. In combination, Anets and Arules provide researchers a powerful tool to create a bird's-eye view of the collected information, to extract hidden biological regularities and to generate hypotheses for further experimental validation. To test this framework we applied it to the analysis of metadata of sequenced prokaryotic genomes from the GenomeOnLine Database (GOLD). The overlapping structure of the data provides a good case study for the proposed framework. The generated Anet revealed a hidden structure in metadata of the prokaryotic organisms with three major clusters representing metadata of pathogens, environmental isolates and plant symbionts. The annotations clustered in each group represented a distinct signature profile of metadata for each group and showed a strong link between phenotypic, genomic and environmental features of the organisms.
Authors:
Minsu Kim
Institutions:
Physics, Emory Univ., Atlanta, Georgia
Title:
Steady state growth of E. coli in low ammonium environment
Abstract:
Ammonium is the preferred nitrogen source for many microorganisms. In medium with low ammonium concentrations, enteric bacteria turn on the Ammonium/methylammonium transporter AmtB, an inner membrane protein that transports NH4+ across the cell membrane against a concentration gradient. In order to study ammonium uptake at low NH4+ concentration at neutral pH, we developed a microfluidic flow chamber that maintains a homogenous nutrient environment during the course of exponential cell growth, even at very low concentration of nutrients. Cell growth can be accurately monitored using time-lapse microscopy. We followed steady state growth down to micro-molar range of NH4+ for the wild type and amtB strains. The mutant exhibited reduced growth at ˜20 M of NH4+ while wild type strain is able to maintain the growth rate below that. Using a combination of quantitative gene expression and flux analysis, we show that E. coli cells maintain the internal ammonia concentrations at a steady level barely above the minimal needed for rapid cell growth in poor ammonium conditions. The robust maintenance is achieved by ultra-sensitive response of AmtB activity, which can be understood in light of a simple integral feedback model of carbon-nitrogen coordination suggested by the characterized molecular interactions, without invoking any molecular cooperativity.
Authors:
Babak Momeni1, Kristen A. Brileya2, Matthew W. Fields2, and Wenying Shou1
Institutions:
(1) Division of Basic Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA; (2) Center for Biofilm Engineering, Montana State Univ., Bozeman
Title:
Compositional stability and spatial patterning driven by ecological interactions in microbial communities
Abstract:
Community structure – the population composition and the pattern of spatial distribution of individuals within a microbial community – is often critical to community function. However, understanding the mechanisms of structure formation in natural communities is hampered by the multitude of cell-cell and cell-environment interactions as well as environmental variability. Here, through simulations and experiments on communities in defined environments, we examined how ecological interactions between two distinct partners impacted community structure. Using a simple mathematical model based on the fitness effects of local cell-cell interactions, we made two predictions. First, we showed analytically that disparate initial population ratios have the potential to converge and thus confer compositional stability only if the interaction is beneficial to at least one partner. Second, only strongly-cooperating partners intermix by appearing successively on top of each other. Both predictions were validated in engineered yeast communities engaging in different metabolic interactions. We then used a more detailed and realistic "diffusion model," tailored to the yeast communities and experimentally validated, to explore conditions that were difficult to achieve experimentally. We found that partner intermixing required spatial localization of large benefits for both partners and was otherwise insensitive to initial conditions and interaction dynamics. Even in simulated communities consisting of several species, most of the strongly-cooperating pairs appeared intermixed. Partner intermixing was also observed in a methane-producing bacteria-archaea community cooperating through redox-coupling. We have thus described the expected composition and patterning of microbial communities when the fitness effects of pair-wise interactions are the major driving force. Analogous to how Gause's competitive exclusion principle has fueled research on mechanisms that generate exceptions to the principle, i.e. the coexistence of species, we hope that our work will create a framework to examine other forces that shape community structure.
Author:
Michele K. Nishiguchi
Institution:
Dept. of Biology, New Mexico State Univ., Las Cruces
Title:
From saltwater to genes and back again: How beneficial bacteria can help decipher the evolutionary mechanisms of symbiosis
Abstract:
The mutualistic association between sepiolid squids (Mollusca: Cephalopoda) and their Vibrio symbionts is an experimentally tractable model to study the evolution of animal-bacterial associations through both wild-caught and experimentally evolved populations. Since Vibrio bacteria are environmentally transmitted to new hosts with every generation, it provides a unique opportunity to resolve how changing environmental conditions may effect bacterial infection, colonization, and persistence in different host species. Vibrio bacteria encounter potentially conflicting selective pressures, competing with one another to colonize the sepiolid light organ, but also competing for resources in the environment outside the squid. Both abiotic and biotic factors contribute to the fitness of individual strains of Vibrio bacteria, but which of these factors are amenable to adaptation and eventually lead to a successful beneficial association has yet to be elucidated. This poster will cover how environmental conditions and host specificity lead to the development of symbiotically adapted Vibrio bacteria, generating new ideas on the evolution of beneficial associations.
 Authors:
Shaneka S. Simmons and Raphael D. Isokpehi
Authors:
Shaneka S. Simmons and Raphael D. Isokpehi
Institution: Center for Bioinformatics & Dept. of Biology, Jackson State Univ., Jackson, Mississippi
Title:
Iron transport gene neighbors of stress-responsive major facilitator in the genome of bioenergy relevant Rhodopseudomonas palustris
Abstract:
Gene clustering properties have been used to predict gene functions based on annotation of its gene neighborhoods, with the assumption that conserved gene sets are proximally arranged along the chromosome, are dependent on each other, and that conserved neighbors with similar functions are preserved between several genomes. In bacterial species occupying highly complex and unstable environments, gene order conservation is shaped by the relative importance of genes for cell survival and by interference from various selective pressures imposed upon genome stability, affecting different genes and operons or putative operons. Furthermore, remnants of gene order conservation can be found as highly conserved clusters across very distantly related taxonomic lineages, suggesting that selective processes maintain gene organization in various genomic regions. Through the use of bioinformatics tools available in the Integrated Microbial Genome (IMG) systems, we identified the gene specific chromosomal cassette for R. palustris major facilitator fused Universal Stress Protein domain based on the presence/absence of genes within the neighbor of organisms containing a similar chromosomal cluster. The UspA domain (Pfam accession number PF00582) has been shown to respond to a variety of stressors including those causing oxidative stress. One Usp gene (RPC_3634 or UniProt Q210L7) encodes a 565 aa protein that in addition to the USP domain contains a carbohydrate transport Major Facilitator Superfamily (MFS_1) domain. The fused gene is present only in the genome of strain BisB18. The Pfam functional annotation of the upstream gene cluster of RPC_3634 also revealed genes encoding transporters of iron. Genes (RPC_3632, RPC_3631, and RPC_3630) were predicted to be in a transcription unit in the BioCyc database. Chromosomal cassette alignment using RPC_3631 as gene of focus was performed to determine other prokaryotes with the iron transport cluster. The Usp domain may mediate co-regulation and co-functionality of neighboring genes that are involved in various biochemical pathways in an effort to combat environmental stress.
Authors:
Adam M. Sullivan1,2, Xiaopeng Zhao1,2, Yasuhiro Suzuki3, Eri Ochiai3, Stephen Crutcher3, Michael A. Gilchrist2,4
Institutions:
(1) Dept. of Mechanical, Aerospace and Biomedical Engineering, Univ. of Tennessee, Knoxville;
(2) National Institute of Mathematical and Biological Synthesis, Univ. of Tennessee, Knoxville;
(3) Dept. of Microbiology, Immunology, and Molecular Genetics, Univ. of Kentucky, Lexington;
(4) Dept. of Ecology and Evolutionary Biology, Univ. of Tennessee, Knoxville
Title:
Evidence of asynchronous growth of Toxoplasma gondii cyst based on data-driven model selection
Abstract:
Toxoplasma gondii establishes a chronic infection by forming cysts preferentially in the brain. This chronic infection is one of the most common parasitic infections in humans and can be reactivated to develop life-threatening toxoplasmic encephalitis in immunocompromised patients. Analyzing the growth mechanism of T. gondii cyst is important for understanding the pathogenesis mechanism. We developed a differential equation framework of cyst growth and employed Akaike Information Criteria (AIC) to determine the growth and removal functions that best describe the data of cyst sizes measured from the brains of chronically infected mice. The AIC results showed that the T. gondii cyst size grows at a constant rate. Correspondingly, the per capita growth rate of the parasite is inversely proportional to the number of parasites within a cyst, indicating asynchronous replication of the parasite. The modeling and analysis framework may provide a useful tool for understanding the pathogenesis of many apicomplexan parasites.
Author:
Yu Tanouchi
Institution:
Dept. Biomedical Engineering, Duke Univ.
Title 1:
A synthetic-biology approach to understanding bacterial programmed death and implications for antibiotic treatment
Abstract:
Programmed death is often associated with a bacterial stress response. This behavior appears paradoxical, as it offers no benefit to the individual. This paradox can be explained if the death is 'altruistic': the sacrifice of some cells can benefit the survivors through release of 'public goods'. However, the conditions where bacterial programmed death becomes advantageous have not been unambiguously demonstrated experimentally. Here, we determined such conditions by engineering tunable, stress-induced altruistic death in the bacterium Escherichia coli. Using a mathematical model, we predicted the existence of an optimal programmed death rate that maximizes population growth under stress. We further predicted that altruistic death could generate the 'Eagle effect', a counter-intuitive phenomenon where bacteria appear to grow better when treated with higher antibiotic concentrations. In support of these modeling insights, we experimentally demonstrated both the optimality in programmed death rate and the Eagle effect using our engineered system. These findings fill a critical conceptual gap in the analysis of the evolution of bacterial programmed death, and have implications for a design of antibiotic treatment.
Author:
Yu Tanouchi
Institution:
Dept. Biomedical Engineering, Duke Univ.
Title 2:
Optimality and robustness in QS-mediated regulation of a costly public-good enzyme
Abstract:
Bacteria secrete a variety of 'public-good' exoproducts into their environment. These exoproducts are typically produced under the control of quorum sensing (QS), a signaling mechanism by which bacteria sense and respond to changes in their density.QS appears to provide an advantageous strategy to regulate these costly yet beneficial exoproducts: it delays production until sufficiently high cell density, when the overall benefit of exoproducts outweighs cost of their production. This notion raises several fundamental questions about QS as a general control strategy adopted by bacteria: How much delay is advantageous? Under what conditions does QS-mediated regulation become advantageous? How does this advantage depend on the kinetic properties of QS? How robust is a given QS system to the stochastic events that occur over bacterial lifecycles? To quantitatively address these questions, we engineered a gene circuit in Escherichia coli to control the synthesis and secretion of a costly but beneficial exoenzyme. We show that exoenzyme production is overall advantageous only if initiated at a sufficiently high density. This property sets the potential advantage for QS-mediated regulation when initial density is low and the growth cycle sufficiently long in comparison to exoenzyme response time. This advantage of QS-mediated regulation is robust to varying initial cell densities and growth durations and is particularly striking when bacteria face uncertainty, such as from stochastic dispersal during their lifecycle. We show, however, that for QS to be optimal, its kinetic properties must be appropriately tuned; this property has implications for antibacterial treatments that target QS.
Authors:
KL Whiteson1, S Meinard2, YW Lim1, R Schmieder1, M Haynes1, D Blake2, D Conrad 3, FL Rohwer1
Institutions:
(1) San Diego State Univ.;
(2) Univ. of California, Irvine;
(3) Dept of Medicine, Univ. of California San Diego
Title: Breath gasses as biomarkers in cystic fibrosis
Abstract:
Long-term microbial infections in the lungs of Cystic Fibrosis (CF) patients are complex, individual, and difficult to correlate with patient condition or response to treatment. Gasses found in the breath of CF patients may enable detection of the presence of specific microbial metabolism in a particular microbial community and disease state. Our goal is to find molecules in breath samples that are specific to microbial metabolism and unique to CF patients. Ultimately, we would like to unveil mechanisms that explain how bacteria persist in the CF lung. This information could be used to diagnose and treat infection specifically.
Using the Gas Chromotography and Mass Spectrometry methods established in the Rowland-Blake lab at UCI, we have analyzed seven triplicate breath samples at approximately monthly timepoints from a cystic fibrosis patient and three gender-matched controls along with a room sample to establish the background. We also took induced sputum samples from the same CF patient, at different timepoints, and sequenced the total viral DNA, microbial DNA and RNA.
We find elevated levels of 2,3-butanedione in the CF patient, which is not present in the three healthy controls. 2,3-butanedione is a fermentation product specific to a subset of bacteria including Enterobacter spp, Streptococcus spp and Klebsiella spp. In a bacterial fuel cell model recently published by the Angenent lab at Cornell University, 2,3-butanediol produced by Enterobacter aerogenes fueled increased growth rates in Pseudomonas aeruginosa. When P. aeruginosa had access to 2,3-butanediol as a carbon source, production of a particular phenazine, pyocyanin, greatly increased. Phenazines are redox active compounds that have been described as antibiotics, however they can also reduce Fe(III) to Fe(II), which P. aeruginosa is equipped to uptake with an Fe(II) specific transporter, FeoB. Phenazines can also act as electron acceptors, enabling anaerobic respiration in the low oxygen conditions of CF sputum. We find hits to genes involved in 2,3-butanedione metabolism and catabolism, along with phenazine production and Fe(II) transport in our CF microbial metagenomes.
The combination of 2,3-butanedione in the CF patient’s breath with the presence of genes encoding enzymes required for butanedione and phenazine metabolism suggest that this CF patient’s lung harbors the same productive bacterial interaction as the bacterial fuel cell model. Access to scarce iron resources and the ability to carry out anaerobic respiration could both give P. aeruginosa a survival advantage in the lungs of a CF patient. Synergism between bacteria which arises in the biogeochemical circumstances unique to an individual CF lung may be an important driver of microbial growth and provide explanations for difficult to diagnose exacerbations in CF patients.
NIMBioS
1122 Volunteer Blvd., Suite 106
University of Tennessee
Knoxville,
TN 37996-3410
PH: (865) 974-9334
FAX: (865) 974-9461
Contact NIMBioS


